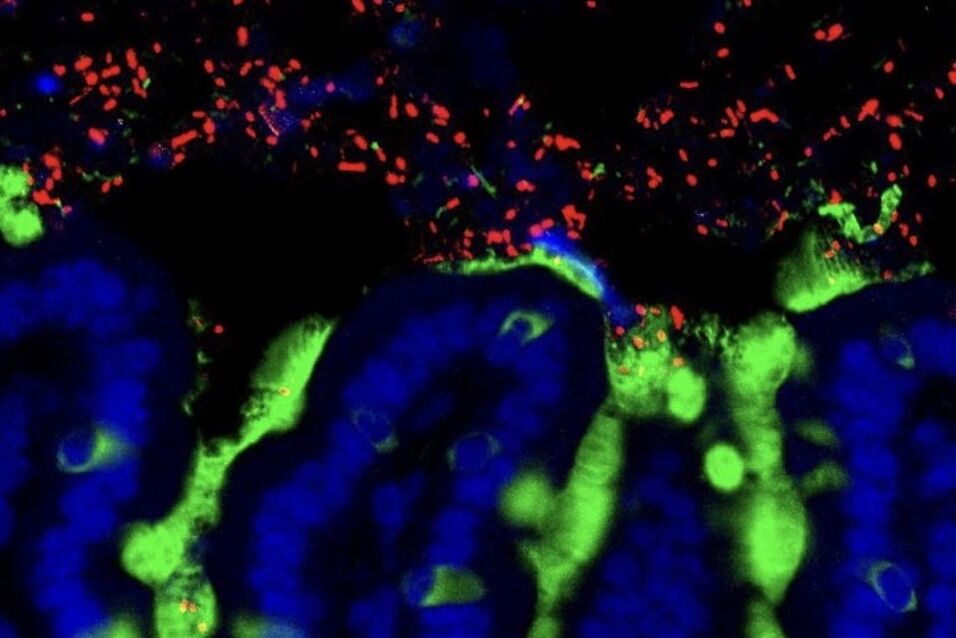
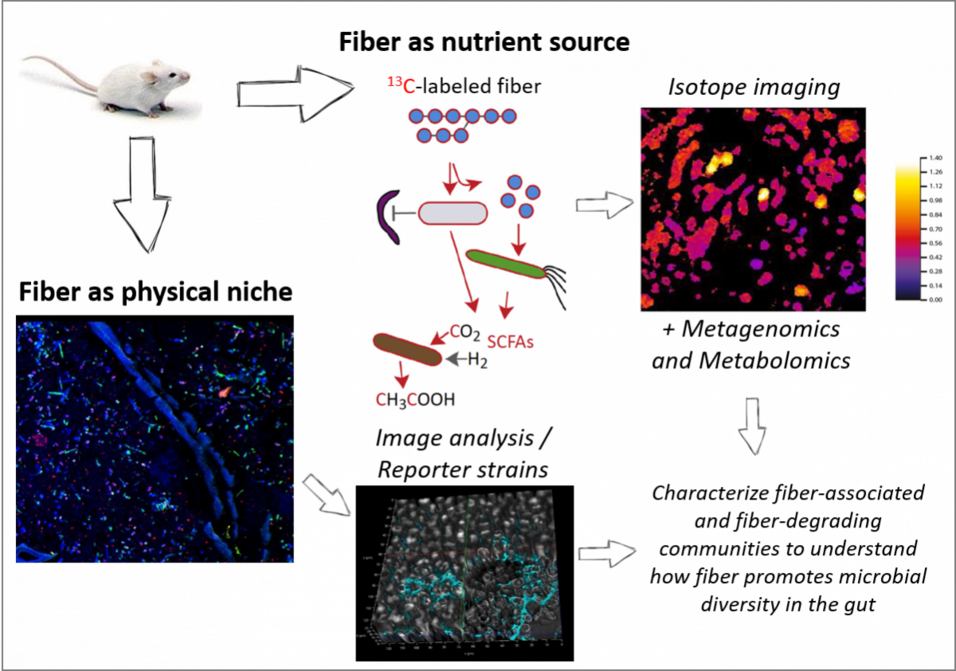
Aims: We aim to elucidate the mechanisms underlying the modification of the gut microbiota by dietary fiber. We will explore two possible, non-mutually-exclusive, mechanisms: (1) that fiber increases microbial diversity by acting as an important nutrient source for the community, and (2) that insoluble fibers increase diversity by offering increased spatial structure, thereby reducing competition by closely-related species. Mechanism 2 is based on observations that closely-related microbial strains can co-inhabit the gut due to differences in physical habitat, which has been formulated as the “Restaurant Hypothesis”. In preliminary research we have observed that fiber-rich diets promote spatial heterogeneity, genetic richness and functional redundancy in the colon microbiota.
Approach: We will use meta-omic as well as single-cell approaches (e.g. FISH) and advanced image analysis (with H. Daims) to detail the fine-scale structuring and activity of the gut microbiota in a mouse model maintained either with or without dietary fiber. We will also employ gene reporter assays in selected model organisms (e.g. Bacteroides thetaiotaomicron, E. coli) introduced into mice as a targeted method to determine how spatial structuring affects microbial activity.
Relevance: This project will give important insights into how diet and dietary component structure shapes the gut microbiota and composition and activity.
Student: Hamid Rasoulimehrabani
Faculty: David Berry (PI), Alexander Loy, Holger Daims
Funding: US DOE Funding
Selected publications:
Pester, M., Maixner, F., Berry, D., Rattei, T., Lücker, S., Richter, A., Spieck, E., Lebedeva, E., Loy, A., Wagner, M., Daims, H. 2014. NxrB encoding the beta subunit of nitrite oxidoreductase as novel functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol. 16(10): 3055-3071. DOI: 10.1111/1462-2920.12300
Daniel, H., Moghaddas Gholami, A., Berry, D., Desmarchelier, C., Hahne, H., Loh, G., Mondot, S., Lepage, P., Rothballer, M., Walker, A., Böhm, C., Wenning, M., Wagner, M., Blaut, M., Schmitt-Kopplin, P., Kuster, B., Haller, D., Clavel, T. 2014. High-fat diet alters gut microbiota physiology in mice. ISME J. 8: 295–308. DOI: 10.1038/ismej.2013.155
Gutierrez, T., Rhodes, G., Mishamandani, S., Berry, D., Whitman, W. B., Nichols, P. D., Semple, K. T., Aitken, M. D. 2014. Polycyclic aromatic hydrocarbon degradation of phytoplankton-associated Arenibacter spp. and description of Arenibacter algicola sp. nov., an aromatic hydrocarbon-degrading bacterium. Appl Environ Microbiol. 80(2): 618-628. DOI: 10.1128/AEM.03104-13.